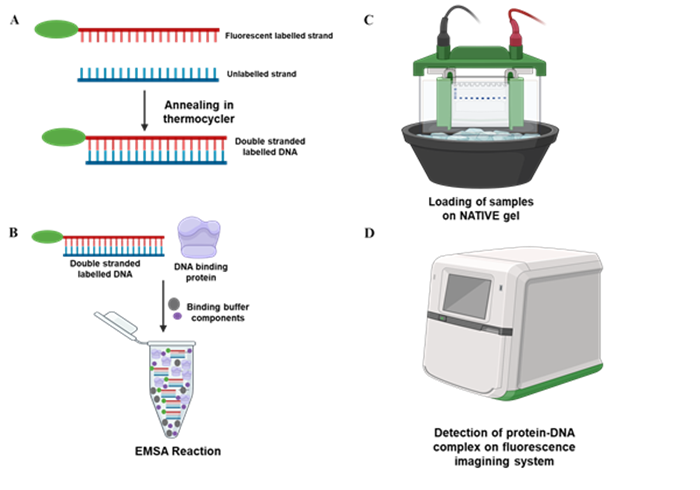
Protein-DNA interaction analysis by electrophoretic mobilty shift assay (EMSA)
The electrophoretic mobility shift assay (EMSA), or gel shift assay is a simple and rapid method to detect protein complexes with nucleic acids. EMSA originally used widely in the study of sequence-specific DNA-binding proteins such as transcription factors, has been further developed to investigate DNA-protein interactions, RNA-protein interactions, and even DNA-RNA interactions. It is also applied to qualify and quantify proteins that specifically bind to given nucleotides, enabling to accommodate a wide range of binding conditions. Mobility-shift assays are often used for qualitative purposes, although under appropriate conditions they can provide quantitative data for the determination of binding stoichiometries, affinities and kinetics.
EMSA based on the principle that the rate of migration of the complex of DNA and protein is slower than single DNA fragment or double stranded oligonucleotide during a nondenaturing polyacrylamide gel. Besides, the kinetic analysis of EMSA is another basic theory to underlying the method. The protein (P), binding to a unique site on a DNA (D), will form a complex PD, in equilibrium with the free components:
$$ P + D \ \underset{kd}{\stackrel{ka}{\rightleftharpoons}} \ PD $$
Where ka is the rate of association and kd is the rate of dissociation. When there is a strong interaction between protein and DNA, Ka>Kd, a distinct band (PD) is observed. However, because of the dissociation occurs during electrophoresis, a faint smear would also show between the two major bands. If a single DNA molecule has multiple binding sites for an individual protein, there will be multiple complexes formed, and we could observe many bands.
Purified proteins, nuclear or cell extract preparations co-incubate with radiolabeled or fluorescent labelled DNA fragment containing the putative protein binding site. Then the reaction products are analysed on a nondenaturing polyacrylamide gel. The specific binding-protein was determined by competition experiments.
EMSA is simple to perform, thus it could be used for a wide range of binding conditions. The assay is also highly sensitive within small protein and nucleic acid concentrations. EMSA is helpful in ranges of nucleic acid sizes and structures. Additionally, the EMSA assay works well for both purified proteins and crude cell extracts.
There are also some limitations of EMSA assay. Firstly, weak interaction or rapid dissociation during electrophoresis can prevent detection of complexes band. Secondly, electrophoretic mobility of a protein-nucleic acid complex depends on many factors other than the size of the protein. So, we cannot measure the protein directly by observing the gel. Thirdly, the result of the assay provides little direct information on the location of the nucleic acid sequences that binding the protein.