



Computation of various physical and chemical properties of Proteins and DNA from their primary sequence
Proteins are one of the most significant and abundant organic molecules in living systems that exhibit more diversity in structure and function than any other classes of macromolecules. They mediate virtually every cellular process exhibiting numerous functions. The diverse functions of proteins are determined by its structure and chemical composition. Although their structures, like their functions, vary greatly, all proteins are made up of one or more chains of amino acids.
Proteins are polymers of amino acids and each amino acid is joined to its neighbour through a covalent amide linkage known as peptide bond. All amino acids share a basic structure, which consists of a central carbon atom, also known as the alpha (α) carbon, bonded to an amino group (NH2 ), a carboxyl group (COOH), and a hydrogen atom. They differ from each other in their side chains or R group, which vary in structure, size, and electric charge and also determine the solubility of the amino acids in water.
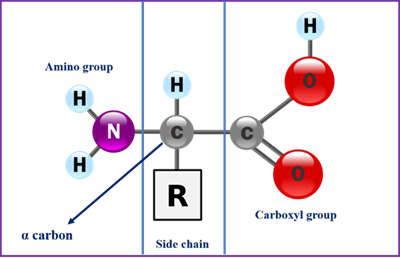
Figure 1: General structure of an amino acid
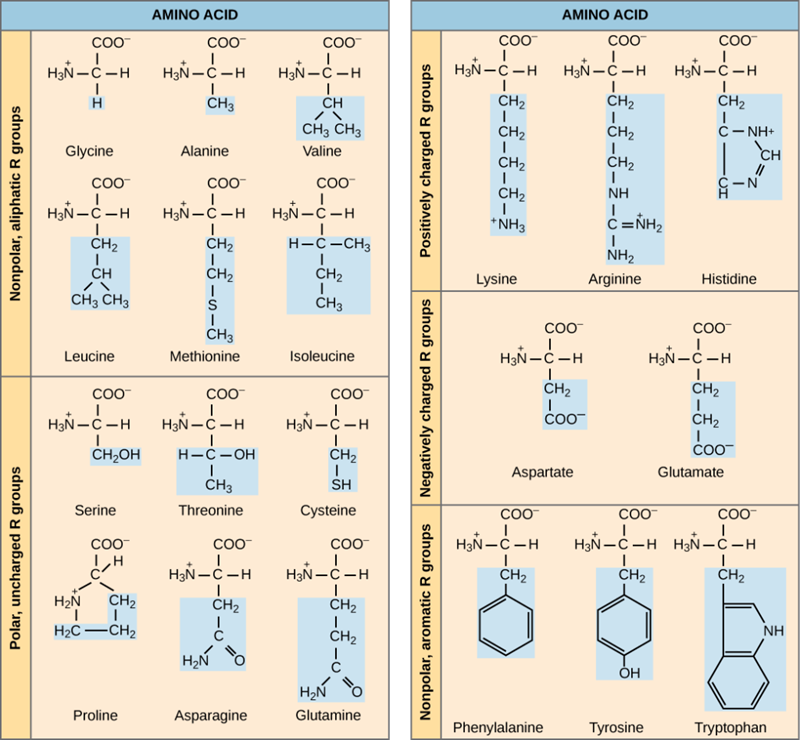
Figure 2: Twenty different amino acids commonly found in proteins, each with a different R group that determines its chemical nature.
The properties of the side chain determine an amino acid’s chemical behavior (that is, whether it is considered acidic, basic, polar, or nonpolar). For example, amino acids such as valine, isoleucine, and leucine are nonpolar and hydrophobic, while amino acids like serine and glutamine have hydrophilic side chains and are polar. Some amino acids, such as lysine and arginine, have side chains that exhibit positively charge at physiological pH and are considered basic amino acids whereas some amino acids like aspartate and glutamate are negatively charged at physiological pH and are considered acidic.
The amino acids of a polypeptide are attached to their neighbours by covalent bonds known as peptide bond. Such a linkage is formed by the removal of elements of water(dehydration) from the α-carboxyl group of one amino acid and the α-amino group of another resulting in a condensation reaction. In a peptide amino acid residue at the end with a free α-amino group is the amino-terminal(or N terminal), the residue at the other end which has a free carboxyl group, is the carboxyl-terminal(or C terminal)
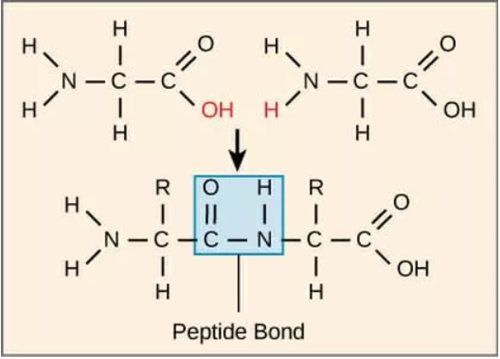
Figure 3 : Peptide bond formed between two amino acids
At wavelength of 280 nm the aromatic amino acids tryptophan (Trp) , tyrosine (Tyr) and phenylalanine(Phe) exhibit strong light absorption and cysteine groups forming disulfide bonds (Cys–Cys) also absorb but to a lesser extent. Consequently, absorption of proteins and peptides at 280 nm is proportional to the content of these amino acids.
DNA(deoxyribonucleic acid) is the molecular repositors of genetic information. Structure and function of every protein, biomolecule and cellular component is dependent on information programmed in the nucleotide sequence of DNA.
A nucleotide has three characteristics component
5 ˈ-phosphate group of one nucleotide is linked to the 3 ˈ hydroxyl group of the next nucleotide, creating a phosphodiester linkage. Each strand of DNA has a backbone made of alternating sugar (deoxyribose) and phosphate groups. Attached to each sugar is one of four bases: adenine (A), cytosine (C), guanine (G) or thymine (T). The two strands are held together by hydrogen bonds between pairs of bases: adenine pairs with thymine, and cytosine pairs with guanine.
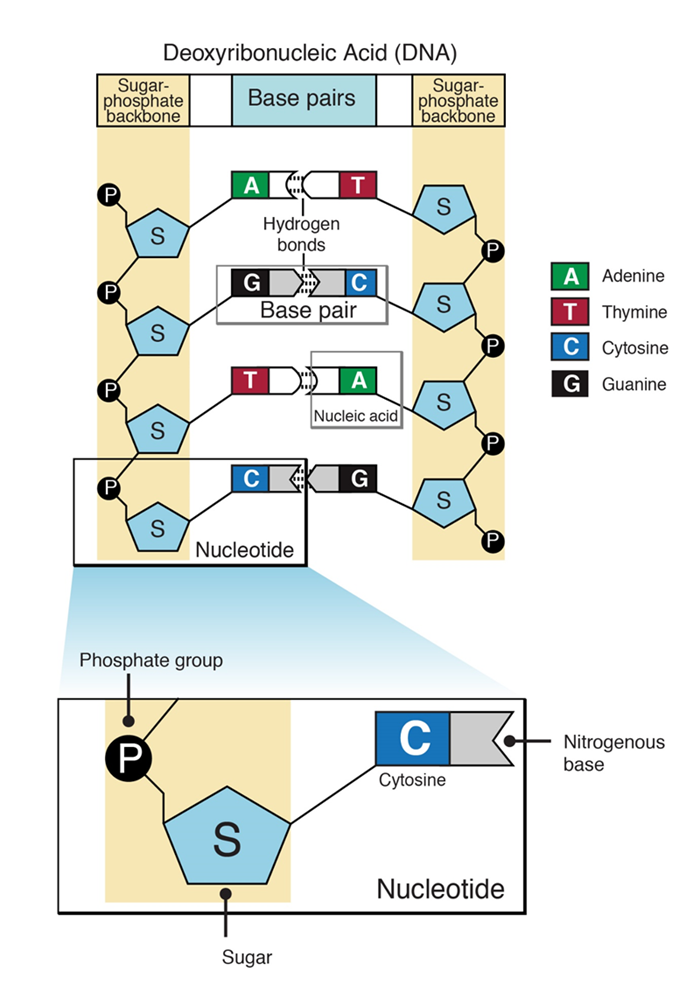
Figure 4 : Schematic representation of structure of a nucleotide and DNA